
This interspecific hybrid population is a cross between colonial bentgrass BCD and creeping bentgrass Providence, grown in a greenhouse. Photo by Jinyoung Y. Barnaby
Colonial bentgrass is a turfgrass species that can be established on grass tennis courts and golf course fairways and used for erosion control (8). Colonial bentgrasses exhibit considerably better resistance to the important fungal disease dollar spot
and have shown improved water-deficit stress tolerance (2). Creeping bentgrass is a fine-textured grass that can tolerate extremely low mowing heights and is widely utilized on golf courses where cool-season grasses are adapted (10). Unfortunately,
creeping bentgrass is susceptible to dollar spot. The fungus is highly active during the spring, summer and early fall in areas that have high humidity, leading to frequent fungicide applications that are costly and have created fungal populations
with fungicide resistance (9). One way to deal with the dollar spot disease issues that exist in the genus Agrostis would be to develop a better understanding of dollar spot resistance in colonial bentgrass and identify plant breeding strategies to
help utilize this resistance.
Hybridization between creeping and colonial bentgrass was first reported by Jones (6), who examined the chromosome pairing of interspecific hybrids and suggested that the species are fairly closely related. Bradshaw (4) studied mixed populations of creeping
and colonial bentgrass and found that little barrier seemed to exist for natural hybridization. More recently, Belanger et al. (3) described dollar spot resistant hybrids between creeping and colonial bentgrass, finding that some hybrids exhibited
essentially no disease symptoms. Warnke (12) utilized DNA markers and high throughput screening techniques to identify creeping and colonial bentgrass plants exhibiting high levels of hybridization.
Previous studies of genetic diversity in colonial bentgrasses and most genetic diversity studies with grass cultivars have scored DNA differences as present or absent to generate a matrix of ones and zeros for each individual by marker combination. This
strategy based on fragment length has limitations, as it reduces the resolution power of the marker system. In addition, sequencing of numerous DNA fragments in outcrossing grasses has shown that many fragments of the same length can have numerous
sequence differences that are not detected by fragment length analysis (personal observation). This creates problems in the analysis, as fragments of the same length are considered identical by descent; however, they often are not. The use of high-resolution
melt (HRM) analysis screens the entire DNA fragment for sequence variation and groups those with the same melt profile into clusters, providing a more accurate assessment of identity by descent.
Considering the potential of creeping × colonial bentgrass interspecific hybrids for both improved dollar spot resistance and enhanced drought tolerance, the objectives of this study were to utilize DNA variation scored using HRM analysis to evaluate
the relationships of cultivated colonial bentgrass varieties relative to a creeping bentgrass cultivar.
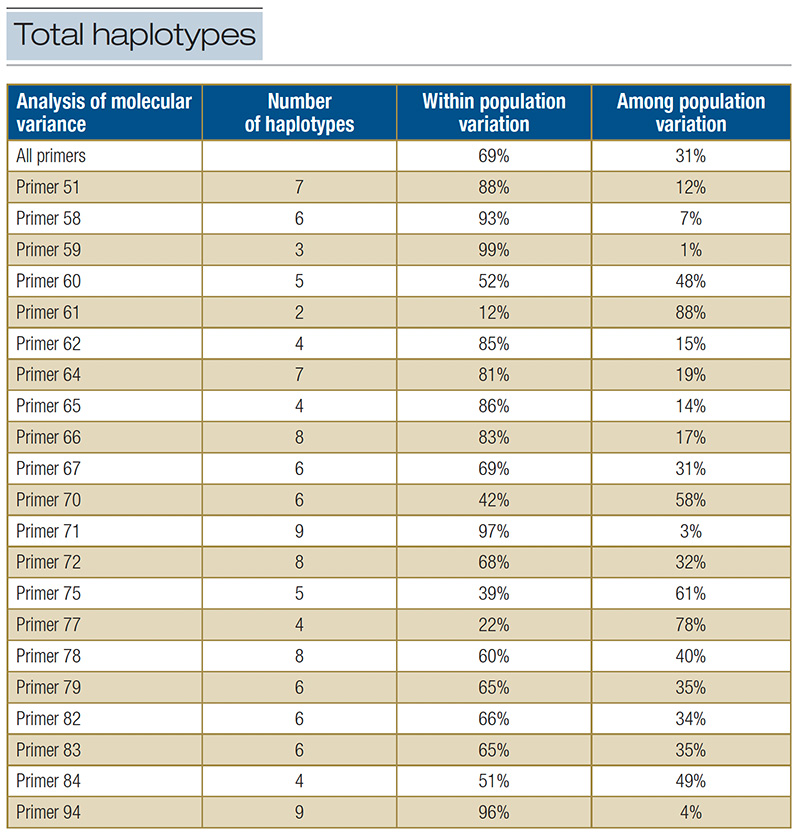
Table 1. The total number of haplotypes calculated by the precision melt analysis software for each SSR primer pair. The
AMOVA was conducted for each primer pair to determine the amount of with and between cultivar variation explained by each.
Materials and methods
A total of 108 colonial bentgrass plants consisting of 12 individual plants from each of the cultivars A08-EBM, PST-R9D7, Greentime, SR7150, BCD, A08-FT12, Greentime2, Tiger2 and Bardot, respectively, and 12 plants of the creeping bentgrass cultivar Tour
Pro were grown in a greenhouse at the USDA-ARS facility in Beltsville, Md. Plants were established in 6-inch (15.2-centimeter) containers in a Pro-Mix potting media. The plants were irrigated to prevent desiccation and maintain consistent plant growth.
The photoperiod was approximately 14 hours throughout the experiment, and greenhouse temperatures ranged from 70 to 82 F (21 to 28 C) during the day and 64 to 73 F (18 to 23 C) at night. Plants were maintained at a 1.97-inch (5-centimeter) canopy
height by regular clipping with scissors. DNA collection was done using FTA (Flinders Technology Associates) cards as described in Warnke (12).
The design of DNA markers used in this study is described in Warnke et al. (11). Forty-eight DNA markers developed from colonial bentgrass were initially screened; 29 amplified and were selected for further testing. High resolution melt analysis was performed
and replicated two times with each plant (Table 1). Distinct melting temperature profiles were scored using computer software that clustered melting temperature profiles, and each unique DNA and melting temperature profile were considered separate
clusters. The most common cluster was labeled cluster 1 and the next most abundant cluster 2 until all clusters were assigned a number. The resulting cluster numbers were used to produce a data matrix representing the DNA variation for each sample.
Each reaction was replicated two times and compared for consistency and fluorescence signal intensity. Genetic distance was calculated with the computer program GENALEX 6.5. DNA variation was separated into within and between population variation
for all DNA markers together and for each marker individually. Principle component analysis was used to visualize the variation of all individuals and all cultivars.
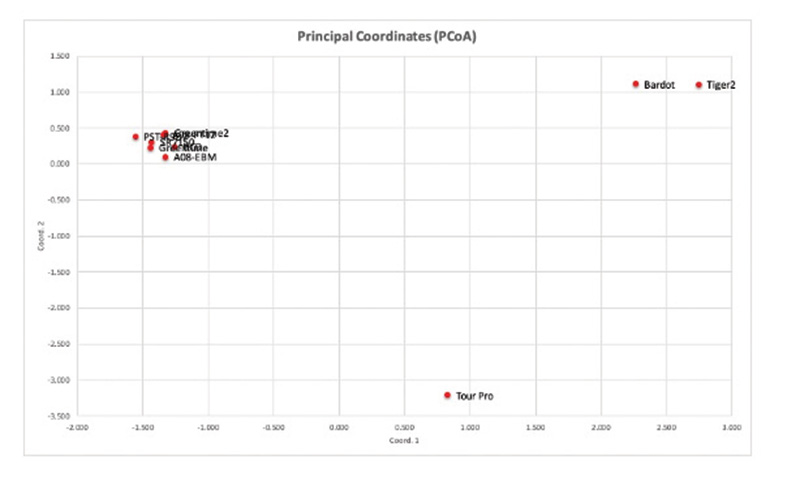
Figure 1. Principal coordinate analysis of the 10 cultivars included in the study. Each cultivar is considered a population in the analysis. The first two principal components explained 93% of the variation.
Results and discussion
The 21 DNA sequences uniquely identified all 120 plants included in the study as would be expected in a highly outcrossing, self-incompatible grass species. DNA variation that is unique in a cultivar are useful for cultivar identification efforts; however,
in this study the limited genetic diversity of colonial bentgrasses only allows the creeping bentgrass Tour Pro and colonial cultivars Tiger 2 and Bardot to be discriminated from the group of colonials consisting of A08-EBM, PST-R9D7, Greentime, SR7150,
BCD and A08-FT12 (Figure 1). The cultivars in this group were very similar genetically; however, it was possible to identify individual plants with similarity to the creeping cultivar and the grouping containing Bardot and Tiger 2 (Figure 2). Considering
the overall sustainability of colonial bentgrasses including the documented high levels of resistance to dollar spot and its potential for hybridization with the more widely utilized dollar spot susceptible species creeping bentgrass, perhaps more
efforts should be put into identifying novel sources of colonial bentgrass, as has been suggested by Belanger et al. (3) and Zhao et al. (13). To further this effort, we have developed a large (300 plants) creeping × colonial bentgrass interspecific
population that we plan to utilize for the identification of genes contributing to important traits such as drought tolerance and dollar spot resistance. The high levels of interspecific hybridization observed previously (12) with some genotypes of
creeping and colonial bentgrass suggests that interspecific hybridization may occur naturally. The data from this study suggests that it could be possible to use DNA variation data to select creeping and colonial bentgrass genotypes with improved
hybridization potential (Figure 2). The presence of alleles that are far more common in the alternate species in plants falling between the species clusters suggests that previous pollinations may have occurred, making these plants good candidates
for plant breeding efforts.
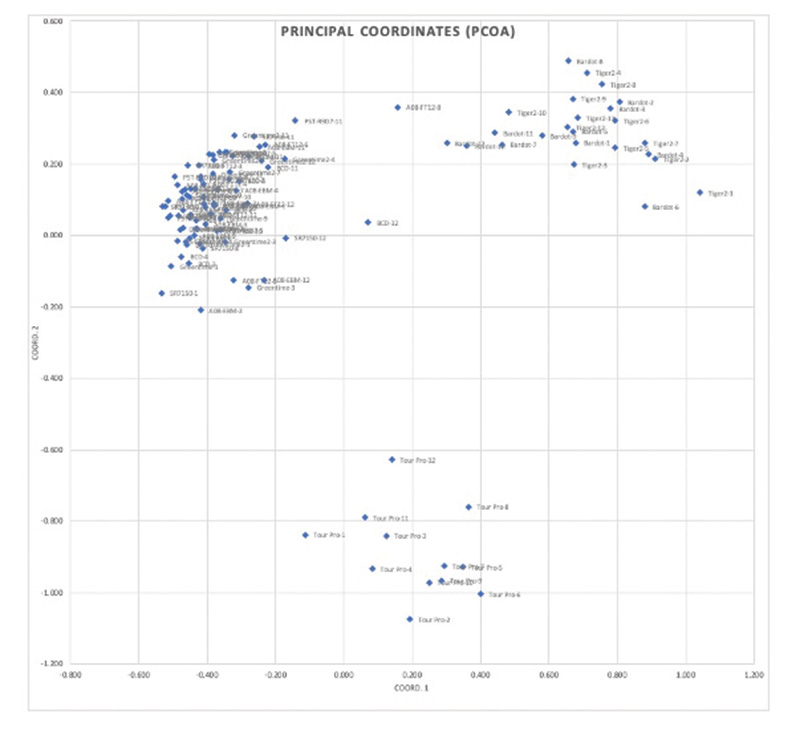
Figure 2. Principal coordinate analysis of the 120 individuals included in the study. The first two principal components explained 43% of the variation.
Both creeping and colonial bentgrass cultivars are developed as populations and are, therefore, variable. Most diversity studies of synthetic cultivars do not consider the levels of genetic variation that exist between individuals within the cultivar.
To estimate this, the mean genetic distance from the genetic distance data of the 12 individuals within each cultivar was calculated (Table 2). The genetic distance values calculated are the sum of the individual DNA differences between two plants
and ranged from a high of 16 differences at the 21 sequences tested in the cultivar Tiger 2 to a low of 1 in the cultivar Greentime (Table 2). Overall, this information shows how genetically different individuals within a synthetic cultivar can be,
and this information should be considered when designing experiments that compare cultivars.
The use of HRM analysis evaluates the sequence of the entire DNA sequence, providing a much more accurate assessment of the similarity of two plants, thus yielding a better estimate of identity among individuals sharing a common ancestor. The use of HRM
analysis to generate DNA variation generally identifies more variation, and this is likely a more accurate assessment of what is occurring in outcrossing grasses. Additionally, HRM analysis is much easier to conduct using commonly available machines
and when combined with rapid DNA extraction allows genetic diversity assessments to be completed much faster and cheaper than studies using other methods.
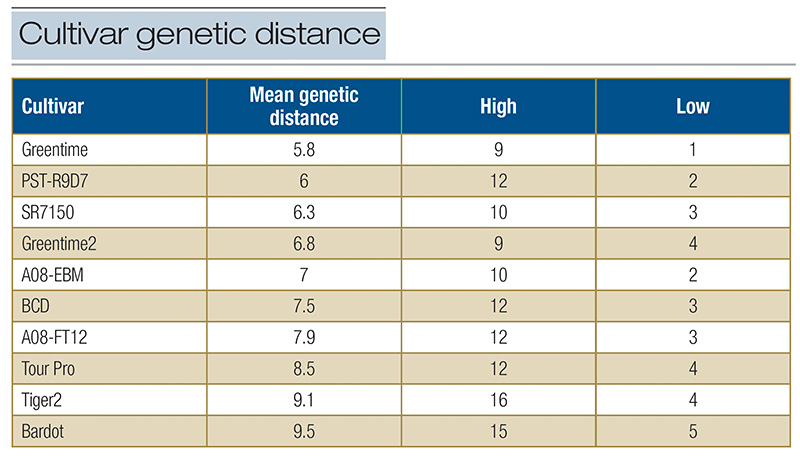
Table 2. The mean within cultivar genetic distance for each cultivar derived by summing the genetic distances between each of the 12 plants in a cultivar and dividing by 66. The high value is the largest distance between any two plants within a cultivar and the low value is the smallest genetic distance between any two plants in a cultivar.
The research says
- The limited genetic diversity of colonial bentgrasses only allows the creeping bentgrass Tour Pro and colonial cultivars Tiger 2 and Bardot to be discriminated from the group of colonials consisting of A08-EBM, PST-R9D7, Greentime, SR7150, BCD and
A08-FT12.
- The data from this study suggests that it could be possible to use DNA variation data to select creeping and colonial bentgrass genotypes with improved hybridization potential.
- Considering the overall sustainability of colonial bentgrasses including the documented high levels of resistance to dollar spot and its potential for hybridization with the more widely utilized dollar spot susceptible species creeping bentgrass,
perhaps more efforts should be put into identifying novel sources of colonial bentgrass.
- Researchers developed a large (300 plants) creeping x colonial bentgrass interspecific population that they plan to utilize for the identification of genes contributing to important traits such as drought tolerance and dollar spot resistance.
Literature cited
- Amundsen, K., and S. Warnke. 2011. Species relationships in the genus Agrostis based on flow cytometry and MITE-display molecular markers. Crop Science 51(3):1224-1231 (https://doi.org/10.2135/cropsci2010.09.0512).
- Amundsen, K., S.E. Warnke, B.S. Bushman, M.D. Robbins, R. Martin and K. Harris-Shultz. 2020. Colonial bentgrass transcript-expression differences compared with creeping bentgrass in response to water-deficit stress. Crop Science 61(3):2135-2147 (https://doi.org/10.1002/csc2.20437).
- Belanger, F.C., S. Bonos and W.A. Meyer. 2004. Dollar spot resistant hybrids between creeping bentgrass and colonial bentgrass. Crop Science 44(2):581-586 (https://doi.org/10.2135/cropsci2004.5810).
- Bradshaw, A.D. 1958. Natural hybridization of Agrostis tenuis Sibth. and A. stolonifera L. New Phytologist 57:66-84.
- Honig, J.A., C. Kubik, V. Averello, J. Vaiciunas, W.A. Meyer and S.A. Bonos. 2016. Classification of bentgrass (Agrostis) cultivars and accessions based on microsatellite (SSR) markers. Genetic Resources and Crop Evolution 63:1139-1160 (https://link.springer.com/article/10.1007/s10722-015-0307-6).
- Jones, K. 1956. Species differentiation in Agrostis II: The significance of chromosome pairing in the tetraploid hybrids of Agrostis canina subsp. montana Hartm., A. capillaris Sibth. and A. stolonifera L. Journal of Genetics 54:377-393.
- Peakall, R., and P.E. Smouse. 2012. GenAIEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research — an update. Bioinformatics 28(19):2537-2539 (https://doi.org/10.1093/bioinformatics/bts460).
- Ruemmele, B.A. 2003. Agrostis capillaris (Agrostis tenuis Sibth) colonial bentgrass. Pages 187-200. In: M.D. Casler and R.R. Duncan, eds. Turfgrass Biology, Genetics, and Breeding. John Wiley & Sons, Hoboken, N.J.
- Sang, H., J.T. Popko Jr. and G. Jung. 2019. Evaluation of a Sclerotinia homoeocarpa population with multiple fungicide resistance phenotypes under differing selection pressures. Plant Disease 103(4):685-690 (https://doi.org/10.1094/PDIS-06-18-1080-RE).
- Warnke, S. 2003. Creeping bentgrass (Agrostis stolonifera L.). Pages 785-790. In: M.D. Casler and R.R. Duncan, eds. Turfgrass Biology, Genetics, and Breeding. John Wiley & Sons, Hoboken, N.J. pp. 785-790.
- Warnke, S.E., C.S. Thammina, K. Amundsen, P. Miljanic and H. Hershman. 2017. High-resolution melt analysis of simple sequence repeats for bentgrass species differentiation. International Turfgrass Society Research Journal 13(1):466-470 (https://doi.org/10.2134/itsrj2016.10.0838).
- Warnke, S.E. 2020. Detection of interspecific hybrids between creeping bentgrass and colonial bentgrass using DNA marker analysis. Agrosystems, Geosciences and Environment 3(1):e20048 (https://doi.org/10.1002/agg2.20048).
- Zhao, H., S.S. Bughrara and J.A. Oliveria. 2006. Genetic diversity in colonial bentgrass (Agrostis capillaris L.) revealed by EcoR1-Mse1 and Pst1-Mse1 AFLP markers. Genome 49:328-335 (https://cdnsciencepub.com/doi/10.1139/g05-113).
Scott E. Warnke (scott.warnke@usda.gov) and Jinyoung Y. Barnaby are research geneticists in the Floral and Nursery Plants Research Unit at the U.S. Department of Agriculture-Agricultural Research Service, U.S. National Arboretum, Beltsville, Md.