
Annual bluegrass (Poa annua L) is listed as one of the top five most common weeds in turfgrass, and within the past three years, it has risen to the most troublesome weed in turfgrass and second most troublesome weed overall, behind only Palmer amaranth
(Amaranthus palmeri) (17). Poa annua has been reported resistant to 12 sites of action (SOA), which equals that of barnyard grass (Echinochloa crus-galli) and is only slightly fewer than that of rigid ryegrass (Lolium rigidum) (8). Across species,
resistance is most common to 5-enolpyruvylshikimate-3 phosphate synthase (EPSPS), acetolactate synthase (ALS), α-tubulin and photosystem II protein D (psbA) inhibitors.
Resistance is a natural, evolved response to human agricultural practices (15), and with over 500 unique instances of herbicide resistance, the ability to identify resistance mechanisms is important for developing alternative management practices (8,
14). Resistance occurs as either target-site resistance (TSR) or non-target-site resistance (NTSR). TSR presents as direct changes to a gene, while NTSR presents as a mechanism related to reduced absorption or translocation of a herbicide or changes
in metabolic interactions (5, 6). While most mechanisms of NTSR are still not very well understood, TSR is more straightforward and is often elucidated through sequencing alone.
In response to the widespread proliferation of P. annua in turfgrass, the ResistPoa Project was formed with the goal of characterizing the distribution of resistant populations and improving management practices, both chemical and nonchemical strategies
(1). The research was conducted to characterize TSR through various gene-sequencing approaches. Populations suspected of resistance based on site history were collected in field conditions and screened for susceptibility to as many as 10 herbicides
representative of nine different SOAs and a single plant growth retardant. Four target sites were selected for sequencing: ALS, EPSPS, psbA and α-tubulin. These four genes were chosen due to the propensity of P. annua resistance to these sites
of action and because there are known TSR mechanisms (2, 9, 11, 12, 13). Given the known herbicide use history of most sampled sites, combined with reports that many collected populations had escaped standard herbicide treatments, it was hypothesized
that most populations would present with TSR mutations.
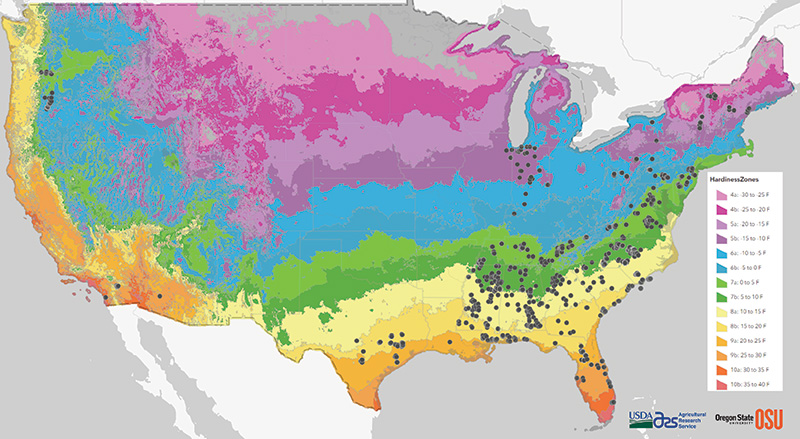
Figure 1. Map of collection sites for all Poa annua populations collected across the USDA plant hardiness zones. Black points indicate a population; however, vicinity of collection sites prevents all 1,367 populations from appearing on the map. USDA plant hardiness zones were delineated using freely reproducible data from the PRISM Climate Group at Oregon State University and USDA-ARS.
Methodology
Collection
Poa annua populations were collected across the United States from areas representing four major turfgrass sectors, including golf courses, home lawns, athletic fields and sod farms. Populations were primarily selected based on herbicide use history and
reports from turfgrass managers that plants had escaped standard herbicide treatment and were suspected to be resistant. To achieve representative sampling of warm-season and cool-season turfgrass across climate gradients, sample collection spanned
multiple USDA plant hardiness zones, with the majority of samples collected between zones 5a and 9b, as depicted in Figure 1 (16).
Approximately 25-30 plants, or about 400 tillers, were collected for each unique population. Coordinates (latitude and longitude) were recorded for each sampling location to facilitate visualization of spatial trends in suspected resistance and target-site
mutations.
Screening
Sampled populations were screened for suspected resistance to as many as 11 unique herbicides (preemergence and postemergence) and plant growth retardants based on regional use patterns and collaborator participation in advanced studies. Herein, we report
on the screening process for herbicides corresponding to the four target sites presented in this study: glyphosate (Round-up, Bayer) (EPSPS inhibitor); foramsulfuron (Revolver, Bayer)/trifloxysulfuron (Monument, Syngenta) (ALS inhibitor); simazine
(Princep, Syngenta) (PS-II inhibitor); and prodiamine (Barricade, Syngenta) and pronamide (Kerb, Corteva Agriscience) (mitotic inhibitor). Postemergence testing was conducted on each collected population by transplanting 20 P. annua tillers into flats
filled with herbicide-free, native soil (indicative of region collection) and established in a greenhouse environment. A total of 10 flats were established for each population and were fertilized and irrigated as needed to prevent stress. Additional
plants were transplanted into separate flats to produce seeds for storage and advancement to subsequent studies as appropriate.
Pertinent information for each herbicide treatment is presented in Table 1. Due to regional limitations in herbicide availability, some sites used trifloxysulfuron to screen for suspected ALS-inhibitor resistance, while others used foramsulfuron. Treatments
were applied one week after transplant using a water carrier volume of 40 gallons per acre and a CO2-pressurized backpack sprayer equipped with 11,004 flat-fan spray nozzles (TeeJet, Spraying Systems Co.). Populations were visually evaluated for injury
21 days after initial treatment (DAT), and populations that survived the standard rate of herbicide treatment were labeled “suspected resistant” and moved forward in the sequencing process.
For preemergence screening, two methods of testing (either seedling emergence or hydroponics) were implemented at the discretion of the cooperators based on local infrastructure and available resources. Because the objective of this screening process
was to identify potential herbicide-resistant plants, the method used for initial preemergence screening was not a major concern. Twenty seeds from each population were placed on the surface of native soil in 4-inch (10-centimeter) pots. Two replicates
of each population were used for screening. Seeds were covered with 0.2 inches (0.5 centimeters) of soil, irrigated and left for two days to allow for imbibement prior to herbicide treatment. Herbicides were mixed and diluted with tap water and applied
to pots using the same CO2-pressurized backpack system described in the postemergence trial. If both replicates emerged at 21 DAT, the population was categorized as suspected resistant. In the hydroponics system, a whole-plant assay was conducted
to ascertain plant response to prodiamine and pronamide at 0.1-1.0 millimolar (4). Populations that did not present with characteristic root clubbing associated with mitotic inhibition were labeled as suspected resistant.
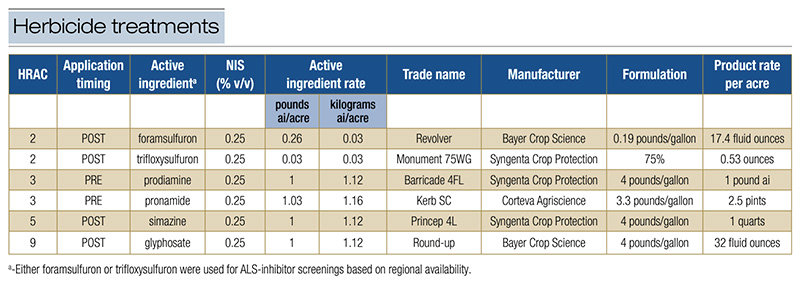
Table 1. Selected herbicide treatments applied to control Poa annua. All rates were standard.
Sequencing
All sequencing results were visually inspected in CLC Genomics Workbench 21 (QIAGEN Sciences) for bulk analysis to be conducted. The susceptible “Auburn” population was mainly utilized in chromatogram comparisons, while the transcriptome data
were used to search for SNPs in the alignments. Chromatograms from capillary sequencing were visually inspected for known SNPs, while FASTQ files produced from AmpSeq were analyzed for SNPs via read mapping and variant calling. Poa infirma and supina
sequences were extracted from the P. annua transcriptome (PRJNA265116, GCZY00000000) and used for subsequent read mapping (3). FASTQ files were trimmed of low-quality reads before read mapping, and each FASTQ file was mapped to both subgenomes for
target-site mutation identification. After read mapping, the basic variant detection was used to identify potential variant sites at a 20% frequency threshold. TSR was considered confirmed if the related SNP was present.
Population attributes including sample identification number, georeferenced sampling location (longitude, latitude), site name, corresponding industry sector (golf course, athletic field, sod production, lawn care operator), preliminary sequencing results
and TSR mutation results for all populations were processed into a standardized format using Excel and exported as comma delimited (.CSV) files. Outputs were subsequently imported into ArcGIS Online (ESRI) and plotted to visualize the spatial distribution
of suspected and confirmed target site mutations. Symbols were used to distinguish sample populations based on specific attributes. USDA plant hardiness zones were delineated using freely reproducible data from the PRISM Climate Group at Oregon State
University and USDA-ARS.
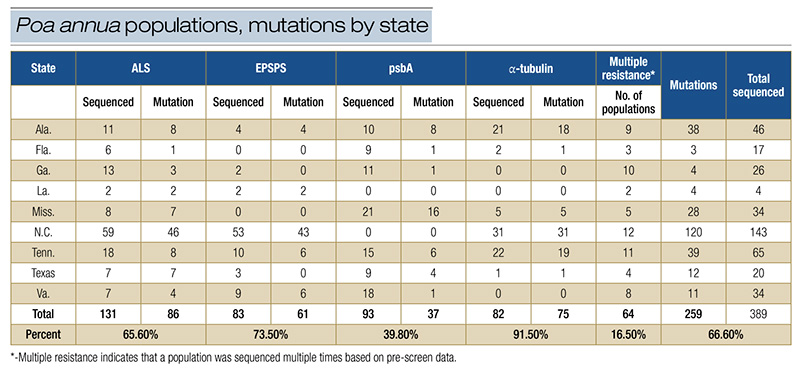
Table 2. Number of total Poa annua populations sequenced by state, percentage of populations found with target-site resistance mutations and without mutations, and percentage of populations with multiple resistance.
Results
Screening
At the end of the collection period, 1,349 populations were collected and screened for resistance. A total of 463 populations passed the initial screening for resistance traits; among these, 389 populations were sequenced. The reduction from passing the
screen to sequencing was due to plants presenting with a resistance type not screened for target-site mutations and plants not surviving transport to Auburn.
Sequencing
No novel mutations were discovered throughout the sequencing process. Single, known mutations were identified for each gene in P. annua conferring resistance to its concomitant SOA. These common mutations were Trp574Leu in ALS, Thr239Ile in α-tubulin,
Pro106Ala in EPSPS, and Ser264Gly in psbA populations. Of the four genes, 65.6% of ALS, 73.5% of EPSPS, 39.8% of psbA, and 91.5% of α-tubulin contained these target-site mutations. The majority of psbA populations presented without a mutation.
Analysis of ALS, EPSPS and α-tubulin populations revealed that more presented a target-site mutation than not. Table 2 describes the number of populations sequenced and whether they contained a target-site mutation.
Discussion
One of the most concerning aspects of this survey was the level of multiple resistance present. After the initial pre-screen, 16.5% of the total populations collected presented with resistance to two or more herbicide families. This included a mix of
populations presenting no mutations for either resistance type, a single mutation corresponding to one resistance type, and a mutation for each resistance type found. The presence of single mutations in populations screened for multiple resistance
indicates the possibility of the co-existence of TSR and NTSR within P. annua populations (10). This level of screened populations with multiple target-site mutations could represent a larger issue for P. annua management in the future.
Implications for management
The ability of P. annua to thrive and evolve surpasses almost every weed species in turfgrass. With resistance to 12 different modes of action, the need to fully understand resistance mechanisms and how to control P. annua seems to present a dire scenario
for practitioners reliant upon chemical control strategies alone. Research presented herein was limited to four general target site mechanisms representative of a large, yet admittedly difficult to quantify, proportion of chemical control strategies
within the turfgrass industry. Mapping (Figure 2) only shows the extent to which resistance is widespread and suggests that regional tactics, albeit important, are not sufficient to circumvent the problem.
Mutations in 66.6% of the 389 populations sequenced across four different SOA were identified by sequencing techniques. Overall, more TSR was confirmed than potential NTSR for all herbicide groups except for PSII inhibitors. Multiple resistance was observed
in 16.5% of the populations sequenced — suggesting an increasingly problematic scenario to P. annua management in turfgrass and one that practitioners and specialists are already anecdotally familiar with.
The origin of resistance is scientifically well understood but is complicated by a poorly understood and as-of-yet measured movement mechanism within turfgrass systems. Undoubtedly, spread is due to human dispersal mechanisms (7, 18). These results further
indicate the tenacity of P. annua as a weed species and the need for alternative management practices.
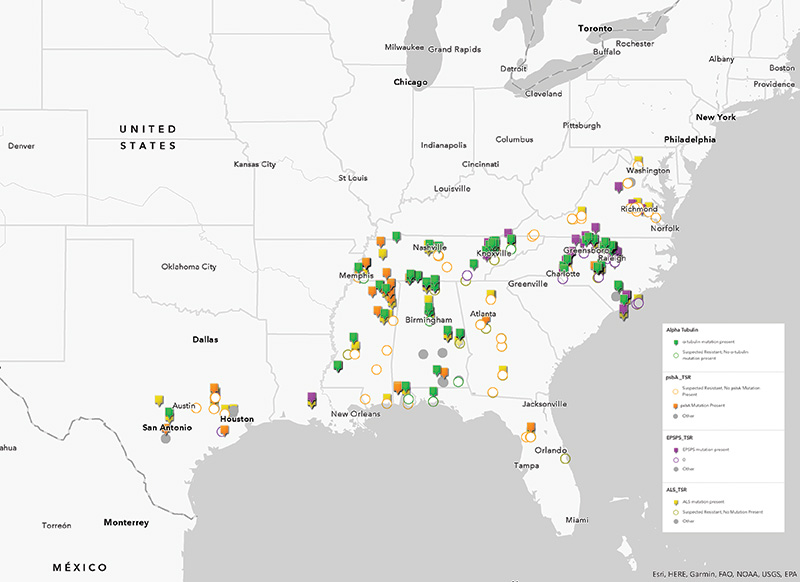
Figure 2. Map depicting suspected resistant populations and their respective resistance type. Points indicate populations with a target-site mutation, while outlined circles indicate populations that screened resistant, but contained no target-site mutation. Gray points indicate populations that screened resistant but were unable to be sequenced for target-site mutations. Yellow points indicate resistance to acetolactate synthase (ALS) inhibitors. Green points indicate resistance to mitotic inhibitors. Purple indicates resistance to 5-enolpyruvylshikimate-3 phosphate synthase (EPSPS) inhibitors. Orange indicates resistance to photosystem II inhibitors.
The research says
- The ability of P. annua to thrive and evolve surpasses almost every weed species in turfgrass, with resistance to 12 different modes of action, presenting a dire scenario for practitioners reliant upon chemical control strategies alone.
- Resistance is widespread and suggests that regional tactics, albeit important, are not sufficient to circumvent the problem.
- Multiple resistance was observed in 16.5% of the populations sequenced — suggesting an increasingly problematic scenario to P. annua management in turfgrass and one that practitioners and specialists are already anecdotally familiar with.
- The origin of resistance is scientifically well understood but is complicated by a poorly understood and as-of-yet measured movement mechanism within turfgrass systems. Undoubtedly, spread is due to human dispersal mechanisms.
Literature cited
- Bagavathiannan, M., J. Brosnan, J. McCurdy, P. McCollough, T. Gannon, B. McCarty, B. Unruh, M. Elmore, A. Patton, S. Askew, J. Kaminski, G. Frisvold, D. Ervin, A. Kowalewski, C. Mattox, B. Grubbs and S. McElroy. 2018. ResistPoa (http://resistpoa.org/).
- Binkholder, K.M., B.S. Fresenburg, T.C. Teuton, X. Xiong and R.J. Smeda. 2011. Selection of glyphosate-resistant annual bluegrass (Poa annua) on a golf course. Weed Science 59(3):286-289 (https://doi.org/10.1614/ws-d-10-00131.1).
- Chen, S., J.S. McElroy, F. Dane and L.R. Goertzen. 2016. Transcriptome assembly and comparison of an allotetraploid weed species, annual bluegrass, with its two diploid progenitor species, Poa supina Schrad and Poa infirma Kunth. Plant Genome 9(1)
(https://doi.org/10.3835/plantgenome2015.06.0050).
- Cutulle, M.A., J.S. McElroy, R.W. Millwood, J.C. Sorochan and C.N. Stewart Jr. 2009. Selection of bioassay method influences detection of annual bluegrass resistance to mitotic-inhibiting herbicides. Crop Science 49(3):1088-1095 (https://doi.org/10.2135/cropsci2008.05.0242).
- Délye, C., M. Jasieniuk and V. Le Corre. 2013. Deciphering the evolution of herbicide resistance in weeds. Trends in Genetics 29(11):649-658 (https://doi.org/10.1016/j.tig.2013.06.001).
- Gaines, T.A., S.O. Duke, S. Morran, C.A.G. Rigon, P.J. Tranel, A. Küpper and F.E. Dayan. 2020. Mechanisms of evolved herbicide resistance. The Journal of Biological Chemistry 295(30):10307-10330 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7383398/).
- Greve, M., and L.R. Pertierra. 2022. Opportunities for studying propagule pressure using gene flow reveal its role in accelerating biological invasions. Molecular Ecology 31(6):1609-1611 (https://onlinelibrary.wiley.com/doi/pdf/10.1111/mec.16385).
- Heap, I. 2022. Resistant species by number of sites of action (top 15). International Herbicide-Resistant Weed Database. Weed Science (https://www.weed science.org/Home.aspx).
- Isgrigg, J., III, F.H. Yelverton, C. Brownie and L.S. Warren. 2002. Dinitroaniline resistant annual bluegrass in North Carolina. Weed Science 50(1):86-90 (https://www.jstor.org/stable/4046445).
- Jugulam, M., and C. Shyam. 2019. Non-target-site resistance to herbicides: Recent developments. Plants (8)10:417 (https://doi.org/10.3390/plants8100417).
- Kelly, S.T., G.E. Coats and D.S. Luthe. 1999. Mode of resistance of triazine-resistant annual bluegrass (Poa annua). Weed Technology 13(4):747-752 (https://doi.org/10.1017/s0890037x00042172).
- McElroy, J.S., M.L. Flessner, Z. Wang, F. Dane, R.H. Walker and G.R. Wehtje. 2013. A Trp574 to Leu amino acid substitution in the ALS gene of annual bluegrass (Poa annua) is associated with resistance to ALS-inhibiting herbicides. Weed Science 61(1):21-25
(https://doi.org/10.1614/ws-d-12-00068.1).
- Mechant, E., T. De Marez, O. Hermann, R. Olsson and R. Bulcke. 2008. Target site resistance to metamitron in Chenopodium album L. Journal of Plant Diseases and Protection 21:37-40 (https://www.researchgate.net/publication/263353421_ Target_site_resistance_to_metamitron_in_
Chenopodium_album_L).
- Norsworthy, J.K., S.M. Ward, D.R. Shaw, R.S. Llewellyn, et al. 2012. Reducing the risks of herbicide resistance: Best management practices and recommendations. Weed Science 60(SP1):31-62 (https://doi.org/10.1614/WS-D-11-00155.1).
- Powles, S.B., and Q. Yu. 2010. Evolution in action: Plants resistant to herbicides. Annual Review of Plant Biology 61:317-347 (https://doi.org/10.1146/annurev-arplant-042809-112119).
- PRISM Climate Group, Oregon State University, https://prism.oregonstate.edu. Data created 2012, accessed May 15, 2020.
- Van Wychen, L. 2020. 2020 survey of the most common and troublesome weeds in grass crops, pasture and turf in the United States and Canada. Online (https://wssa.net/wp-content/uploads/2020-Weed-Survey_grass-crops.xlsx).
- Wódkiewicz, M., K.J. Chwedorzewska, P.T. Bednarek, A. Znój, P. Androsiuk and H. Galera. 2018. How much of the invader’s genetic variability can slip between our fingers? A case study of secondary dispersal of Poa annua on King
George Island (Antarctica). Ecology and Evolution 8(1):592-600 (https://doi.org/10.1002/ece3.3675).
Acknowledgements
This research was supported by the Alabama Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. This project was partially funded by an USDA-NIFA Specialty Crops Research
Initiative (SCRI) program (award No. 2018-51181-28436).
Claudia Ann Rutland (car0053@auburn.edu) is a doctoral student, Nathan D. Hall is a graduate teaching assistant, Jinesh Patel is a research assistant, and Scott McElroy, Ph.D., is a professor of weed science and turf, all at Auburn University, Auburn,
Ala.
Shawn D. Askew, Ph.D., is a professor in the School of Plant and Environmental Sciences, and Eli C. Russell is a doctoral student, both at Virginia Tech, Blacksburg;
Muthukumar V. Bagavathiannan, Ph.D., is an assistant professor, and Daniel Hathcoat is a research specialist, both at Texas A&M University, College Station.
James T. Brosnan, Ph.D., is a professor and director of the Weed Diagnostics Center, and Rebecca G. Bowling, Ph.D., is a turfgrass Extension specialist, both at
the University of Tennessee, Knoxville.
Lambert B. McCarty, Ph.D., is a professor of plant and environmental sciences at Clemson University, Clemson, S.C..
Aaron J. Patton, Ph.D., is a professor of horticulture at Purdue University, West Lafayette, Ind.
Travis W. Gannon, Ph.D., is an associate professor of pesticide fate and behavior at North Carolina State University, Raleigh.
Clebson Gonçalves, Ph.D., is a diversified agricultural adviser at the University of California, Lakeport.
Patrick E. McCullough is a professor of crop and soil sciences at the University of Georgia, Griffin.
Jay McCurdy, Ph.D., is an associate professor in the Department of Plant and Soil Sciences at Mississippi State University.
J. Bryan Unruh, Ph.D., is a professor and associate center director at the University of Florida, Institute of Food and Agricultural Sciences’ West Florida Research and Education Center in Jay, Fla.