
Routine turfgrass clippings collected from a National Turfgrass Evaluation Program (NTEP) trial in College Park, Md., were used to quantify Clarireedia jacksonii DNA using droplet digital polymerase chain reaction (ddPCR). Photo by Krishna Ghimire
Dollar spot, caused by the fungal pathogen Clarireedia jacksonii, remains one of the most economically damaging diseases of cool-season turfgrass in the U.S. Golf courses and sports facilities collectively spend an estimated $500 million annually managing this disease, largely through repeated fungicide applications. On many sites, 10 or more sprays are applied each season, often according to a fixed calendar schedule rather than in response to measured disease pressure.
This management strategy reflects a longstanding limitation in dollar spot control: Infection begins well before visible symptoms develop. By the time characteristic bleached patches appear, the pathogen is already established in the turf canopy. Because early infection cannot be readily detected through visual scouting, fungicides are commonly applied preventively as insurance against outbreaks. While this approach protects turf quality and playability, it also increases costs, labor demands, chemical inputs and concern over fungicide resistance.
Despite decades of research, turf managers have lacked a practical way to determine when the dollar spot pathogen is increasing under field conditions. Recent advances in molecular diagnostics have begun to address this gap, and new work by researchers at the U.S. Department of Agriculture-Agricultural Research Service in Beltsville, Md., builds on these efforts by providing a highly sensitive, field-applicable approach to pathogen monitoring.
Building on earlier molecular detection efforts
The application of DNA-based tools to dollar spot is not entirely new. Researchers at Rutgers University previously developed a quantitative PCR (qPCR) assay for detecting Clarireedia species in turfgrass tissue (1). Their work showed that pathogen DNA could be detected in both symptomatic and asymptomatic creeping bentgrass leaves, providing an important proof of concept that molecular detection can reveal infection before symptoms become obvious.
That study focused primarily on pathogen detection and assay development, using a single cultivar to demonstrate sensitivity and feasibility. It helped establish that molecular approaches could complement visual scouting. However, several practical and biological questions remained unresolved for turf management and breeding:
- Can pathogen DNA be reliably quantified from routine field clippings?
- Can molecular measurements differentiate among cultivars with known differences in dollar spot resistance?
- Can tracking pathogen levels over time provide insight into disease progression and epidemic patterns under natural infection?
The work described here was designed to address these questions while acknowledging and building upon earlier molecular advances.
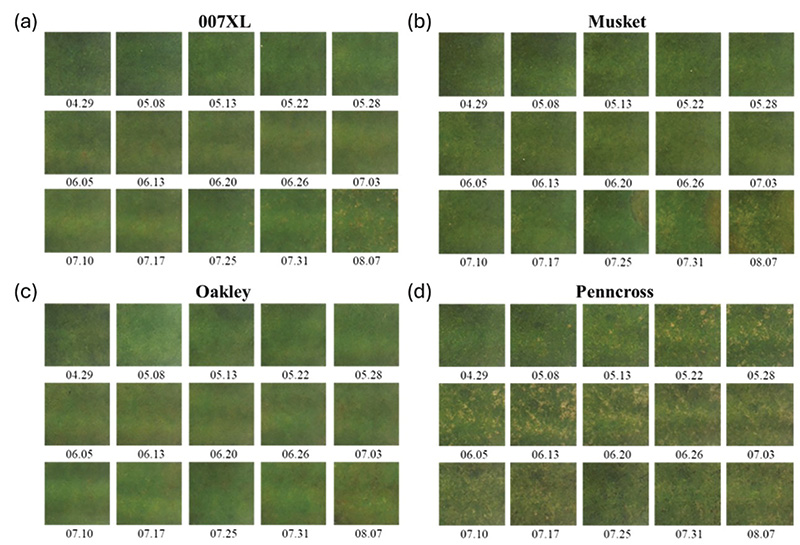
Figure 1. Grid of weekly field plot images showing disease progression in four bentgrass cultivars. — colonial bentgrass: Musket (b); and creeping bentgrass: 007XL (a), Oakley (c), and Penncross (d).
An early-warning DNA test using droplet digital PCR
USDA-ARS researchers in Beltsville developed an ultra-sensitive droplet digital PCR (ddPCR) assay to quantify Clarireedia jacksonii in field-collected turfgrass clippings, enabling comparisons of dollar spot disease dynamics among four bentgrass cultivars displaying differences in resistance to dollar spot. This work was published on July 9, 2025, in Crop Science (2). While qPCR estimates DNA concentration relative to a standard curve, ddPCR partitions each sample into thousands of microscopic droplets and counts the number containing the target DNA. This enables absolute quantification of pathogen DNA and reduces variability associated with amplification efficiency, standard curves and PCR inhibitors commonly found in field-derived plant samples. Using custom primers and probes developed in-house, the assay was optimized specifically for C. jacksonii and validated for use with routine turfgrass clippings.
A key advantage of this approach is practicality. The assay works on routine mowing clippings, requiring no special sampling or destructive collection. Clippings that would normally be discarded can be used to extract DNA and quantify pathogen levels. The sensitivity of the assay allows detection of trace amounts of fungal DNA, providing a highly sensitive early warning indicator of increasing disease pressure — often days to weeks, and in some cases months, before visible symptoms appear.
Measuring disease development under real field conditions
To evaluate the biological and practical value of this early warning system, the ddPCR assay was applied to turfgrass clippings collected weekly from a National Turfgrass Evaluation Program (NTEP) fairway trial in Maryland. Four bentgrass cultivars with established differences in dollar spot susceptibility were included: three creeping bentgrass cultivars and one colonial bentgrass cultivar.
Figure 1 illustrates weekly visual progression of dollar spot symptoms across four bentgrass cultivars under field conditions. Early in the growing season, visible symptoms were minimal across all cultivars, despite known differences in susceptibility. As the season progressed, disease symptoms emerged at different times and intensities, consistent with cultivar resistance levels. The susceptible cultivar Penncross exhibited earlier symptom onset and more extensive disease development, whereas Oakley and 007XL showed delayed and less-severe symptom expression. Across all cultivars, symptom development appeared abrupt, with disease severity increasing rapidly once visible symptoms became established.
Figure 2 compares weekly visual disease severity with ddPCR-based quantification of Clarireedia jacksonii DNA. In all cultivars, increases in pathogen DNA were detected before corresponding increases in visual disease severity. In Oakley and 007XL, pathogen DNA accumulated gradually and remained low for much of the season, often without accompanying visible symptoms. In contrast, Penncross exhibited higher DNA levels earlier in the season, followed by pronounced increases in disease severity. Musket showed an intermediate pattern, with pathogen DNA increases preceding visible symptom development by several sampling dates.
This temporal gap helps explain why dollar spot outbreaks can appear suddenly, even when turf seemed healthy only days earlier. The pathogen has already reached damaging levels by the time symptoms become visible.
Genotypic differences in pathogen dynamics
One of the most distinctive contributions of this work is the demonstration that pathogen DNA dynamics differ among turfgrass genotypes. Cultivars with higher dollar spot resistance maintained low pathogen levels early in the season, delaying symptom development and limiting early epidemic progression. In contrast, the susceptible cultivar exhibited higher early season pathogen levels and experienced two distinct epidemic waves during the growing season. These repeated increases in pathogen DNA were closely associated with increased disease severity. Importantly, not all resistant cultivars behaved in the same way. Some appeared to suppress early pathogen establishment, while others tolerated relatively higher pathogen levels before symptoms became apparent. These patterns suggest that resistance may involve both suppression and tolerance mechanisms, rather than a single uniform response. This finding clarifies what resistance looks like in the field. Resistance is not simply the absence of symptoms; it reflects underlying biological differences in how turfgrass cultivars interact with the pathogen over time. DNA-based quantification allows these differences to be measured directly rather than inferred from visual ratings alone.
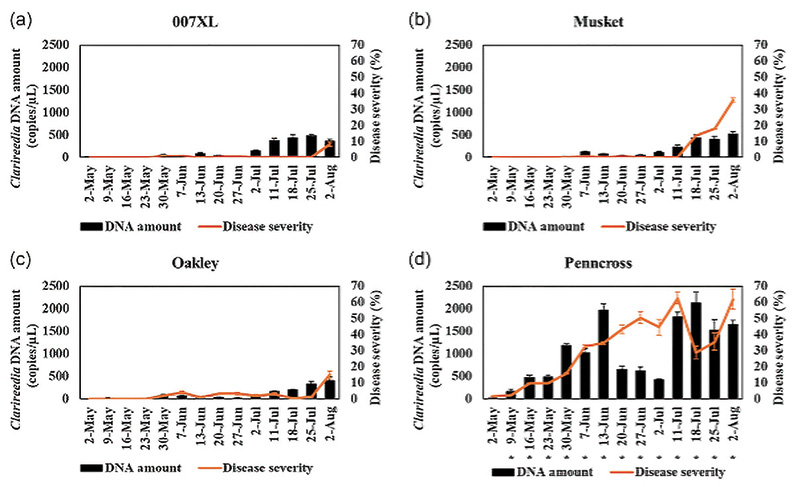
Figure 2. Weekly measurement of Clarireedia jacksonii DNA quantity (copies/µL; black bars) and disease severity
Implications for fungicide timing
The most immediate management implication of this work is improved fungicide timing. Because visual scouting detects dollar spot only after infection has progressed, calendar-based preventive programs remain common. The ddPCR assay provides additional information by identifying when pathogen levels begin to rise, offering an objective indicator of increasing disease risk. Rather than replacing fungicides or traditional scouting, DNA-based monitoring can complement existing decision-making tools. When pathogen DNA remains low, fungicide applications may be delayed with greater confidence. When DNA levels increase, treatments can be timed more precisely to coincide with early disease development. Based on disease progression patterns observed in this study, integrating pathogen detection into management programs has the potential to reduce fungicide applications and costs by an estimated 20%-50%, depending on cultivar susceptibility, site conditions and management objectives. These reductions reflect decreased reliance on routine calendar sprays rather than elimination of fungicide use. Fungicides remain essential for maintaining high-quality turf. The value of early detection lies in using them more efficiently, applying products when they are most likely to provide benefit.
Value for turfgrass breeding and evaluation
The ddPCR assay also offers significant benefits for turfgrass breeding and cultivar evaluation. Traditional resistance assessments rely heavily on visual disease ratings, which can be influenced by environmental conditions, timing and observer variability. DNA-based quantification provides an objective, repeatable measure of pathogen load, even when visual symptoms are minimal or absent.
This capability allows breeders to:
- Identify cultivars that suppress early pathogen establishment.
- Distinguish tolerance from pathogen suppression responses.
- Evaluate resistance under natural infection conditions.
By linking pathogen dynamics directly to cultivar performance, breeders gain clearer insight into resistance mechanisms and can make more informed selection decisions. Over time, this approach supports the development of turfgrass varieties that naturally require fewer fungicide inputs under comparable management conditions.
Economic and environmental considerations
Dollar spot management represents one of the largest recurring expenses in turfgrass systems. Even modest reductions in fungicide use can translate into meaningful savings at the course level. When considered across the industry, the cumulative economic impact is substantial. Reducing unnecessary applications also lowers chemical load, decreases labor and fuel use and reduces selection pressure for fungicide resistance. These outcomes align with increasing expectations for responsible pesticide use while maintaining high standards for turf quality and playability.
Current limitations and future directions
At present, ddPCR is best suited for research applications, breeding trials and diagnostic support rather than routine, on-site testing. However, as molecular technologies become more accessible and streamlined, their integration into field-level decision support may expand. Future research may refine DNA thresholds that signal increased disease risk, explore integration with weather-based risk models and evaluate similar approaches for other turfgrass diseases. Continued work will also help clarify how suppression and tolerance mechanisms vary among cultivars and environments.
Conclusion
This study demonstrates that direct measurement of the dollar spot pathogen provides insight into disease development that is not available through visual scouting alone. By detecting increases in Clarireedia jacksonii DNA before symptoms appear, the ddPCR assay offers a practical early warning indicator of disease pressure under field conditions. Used alongside existing management tools, DNA-based monitoring can improve fungicide timing, support more accurate resistance evaluation and inform breeding efforts aimed at reducing chemical inputs. While not a stand-alone solution, this approach represents a meaningful step toward more precise, informed and sustainable dollar spot management in turfgrass systems.
The research says
- This study demonstrates that direct measurement of the dollar spot pathogen provides insight into disease development that is not available through visual scouting alone.
- By detecting increases in Clarireedia jacksonii DNA before symptoms appear, the ddPCR assay offers a practical early-warning indicator of disease pressure under field conditions.
- Used alongside existing management tools, DNA-based monitoring can improve fungicide timing, support more accurate resistance evaluation and inform breeding efforts aimed at reducing chemical inputs.
- While not a stand-alone solution, this approach represents a meaningful step toward more precise, informed and sustainable dollar spot management in turfgrass systems.
Literature cited
- Groben G., B.B. Clarke, J. Murphy, P. Koch, J.A. Crouch, S. Lee and N. Zhang. 2020. Real-time PCR detection of Clarireedia spp., the causal agents of dollar spot in turfgrasses. Plant Disease 104(12):3118-3123 (https://doi.org/10.1094/PDIS-04-20-0726-RE).
- Ghimire K., Y.H. Kim, J.Y. Barnaby and S.E. Warnke. 2025. Quantification of dollar spot inoculum (Clarireedia spp.) in bentgrass clippings using droplet digital polymerase chain reaction (ddPCR). Crop Science 65(4):e70121 (https://doi.org/10.1002/csc2.70121).
Jinyoung Y. Barnaby (Jinyoung.barnaby@usda.gov) and Scott E. Warnke (retired) are research geneticists in the Floral and Nursery Plants Research Unit at the U.S. Department of Agriculture-Agricultural Research Service, U.S. National Arboretum, Beltsville, Md.